New insights on medulloblastoma from single-cell sequencing

Medulloblastoma, a malignant tumor in the cerebellum, is one of the most common malignant brain cancers in children. Survival rates range from 20 to 90 percent, depending on the genetic subtype. There are at least four: WNT and Sonic Hedgehog (named for the signaling pathways that drive them), Group 3, and Group 4. Group 3 and Group 4, accounting for 60 percent of all medulloblastomas, are the least understood. They often have poor outcomes; many are metastatic at the time of diagnosis.
Existing therapies for medulloblastoma can be toxic and can lead to impaired neurocognitive function and secondary cancers. Targeted therapeutics have been difficult to develop, since some of the genetic pathways involved in the different subtypes are also active in non-cancerous cells.
Seeking better treatment targets, past studies have looked at bulk tumor DNA, RNA, and proteomics. A new study in the August issue of Nature is the first to apply single-cell RNA sequencing, sampling many hundreds of cells from each medulloblastoma subtype — in all, more than 8,700 individual tumor cells from 25 patients. This readout of what genes each cell was turning on and off sheds new light on how the different subtypes originate during the development of the cerebellum and how they relate to each other biologically.
Medulloblastoma and early cerebellar development
Medulloblastomas, like other brain cancers in children, can be seen as cells that have arisen from normally developing brain cells, evolving into genetically distinct malignant populations.
“With single-cell RNA sequencing, we were able to see that medulloblastomas recapitulate parts of normal cerebellar development,” says Mariella Filbin, MD, PhD, a pediatric neuro-oncologist at the Dana-Farber/Boston Children’s Cancer and Blood Disorders Center and a co-first author on the study. “We found that medulloblastoma subtypes are comprised of undifferentiated stem-like cancer cells and differentiating cancer cells.”
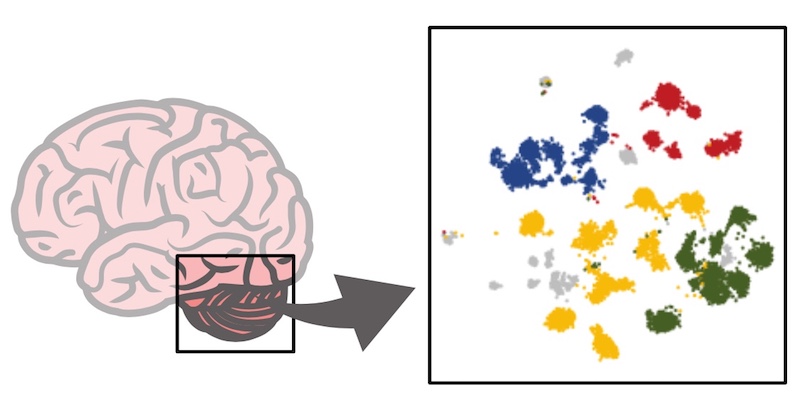
The study showed that Group 3 and Group 4 tumor subtypes shared features with developing brain-cell hierarchies, with Group 3 tumors dominated by primitive progenitor-like cells and Group 4 tumors containing more mature neuronal cells. The study also revealed an intermediate type of medulloblastoma straddling Group 3 and Group 4, consisting of a mix of immature and mature tumor cells.
“For the first time we were also able to propose the cells of origin for group 4 medulloblastoma: unipolar brush cells and glutamatergic cerebellar nuclei cells of the cerebellum,” says Filbin. “For the Sonic Hedgehog subtype, we confirmed cerebellar granule cell precursor cells as the cells of origin.”
While the study did not pin down a cell of origin for Group 3 or WNT tumors, the results raise two possibilities. One is that these tumors arise from tissues outside the cerebellum. Alternatively, the MYC oncogene, which characterizes Group 3, may profoundly alter the cells of origin, making them unrecognizable.
New therapeutic approaches?

Single-cell RNA sequencing is paying great dividends in other brain tumors, like DIPG and glioblastoma. The new observations are a first step toward developing trials to test approaches aimed at encouraging medulloblastoma cells to differentiate into less aggressive forms, the researchers say.
Filbin shares first authorship on the paper with Volker Hovestadt, of Massachusetts General Hospital (MGH) and the Broad Institute of Harvard and MIT; and Kyle S. Smith and Laure Bihannic, of St. Jude Children’s Research Hospital. The study’s co-senior authors were Bradley E. Bernstein and Mario L. Suvà, of MGH and the Broad Institute, and Paul A. Northcott of St. Jude. See the paper for a full list of authors and funders.
Related Posts :
-

A new druggable cancer target: RNA-binding proteins on the cell surface
In 2021, research led by Ryan Flynn, MD, PhD, and his mentor, Nobel laureate Carolyn Bertozzi, PhD, opened a new chapter ...
-

Forecasting the future for childhood cancer survivors
Children are much more likely to survive cancer today than 50 years ago. Unfortunately, as adults, many of them develop cardiovascular ...
-

Pediatric high-grade gliomas: Research reveals effective targeting with avapritinib
Pediatric high-grade gliomas, particularly H3K27M diffuse midline gliomas (DMG), are aggressive malignant brain tumors with a poor prognosis. ...
-

Blood across our lifetimes: An age-specific ‘atlas’ tells a dynamic story
The stem cells that form our blood, also known as hematopoietic stem cells (HSCs), are with us throughout our lives. ...