New technique images whole brains with incredible resolution
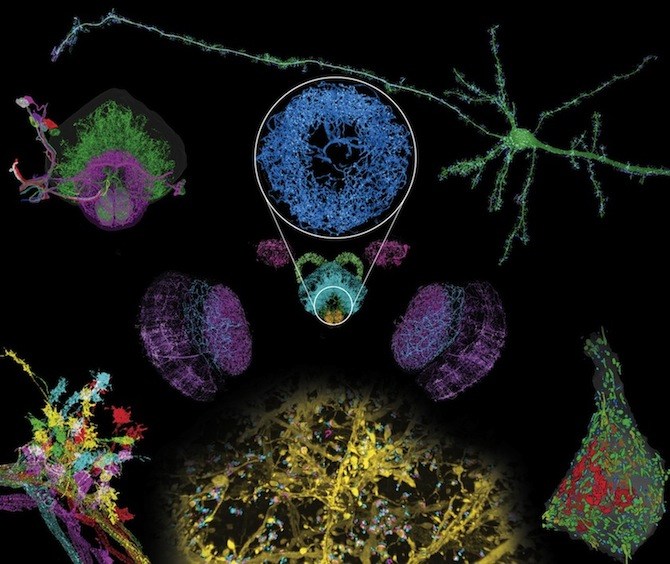
Decades ago, discoveries about the brain’s intricate anatomy were made with careful dissection and drawings. Today, they’re made with super-resolution imaging and massive computing power capable of handling hundreds of terabytes of data.
In this week’s Science, a team out of the Massachusetts Institute of Technology (MIT), the Janelia Research Campus of the Howard Hughes Medical Institute (HHMI), Harvard Medical School (HMS) and Boston Children’s Hospital, describes a technique capable of imaging whole brains at exquisitely high resolution, allowing scientists to distinguish tiny sub-cellular structures.
As co-first author Srigokul Upadhyayula, PhD, of HMS and Boston Children’s explains, “It’s like being able to see the universe and everything in it — the galaxies and the planets — all at the same time, and explore their chemical composition.”
It’s like being able to see the universe and everything in it — the galaxies and the planets — all at the same time.
So far, the team, led by HHMI investigators Eric Betzig, PhD, of Janelia and Ed Boyden, PhD, of MIT, has used the technology to image a complete fly brain and columns of the mouse brain, capturing images in a matter of days that would otherwise take decades with traditional technology. Upadhyayula believes the potential applications for exploring the brain are limited only by the imagination.
How to map a brain
Currently available imaging techniques have led to countless discoveries, but lack the versatility the new method provides. “It was either resolution or volume, you couldn’t have both until now,” explains Upadhyayula, a member of Boston Children’s Program in Cellular and Molecular Medicine. “For the first time, for example, we were able to count all the synapses and map their locations in 3D within a fly brain — over 40 million of them.”
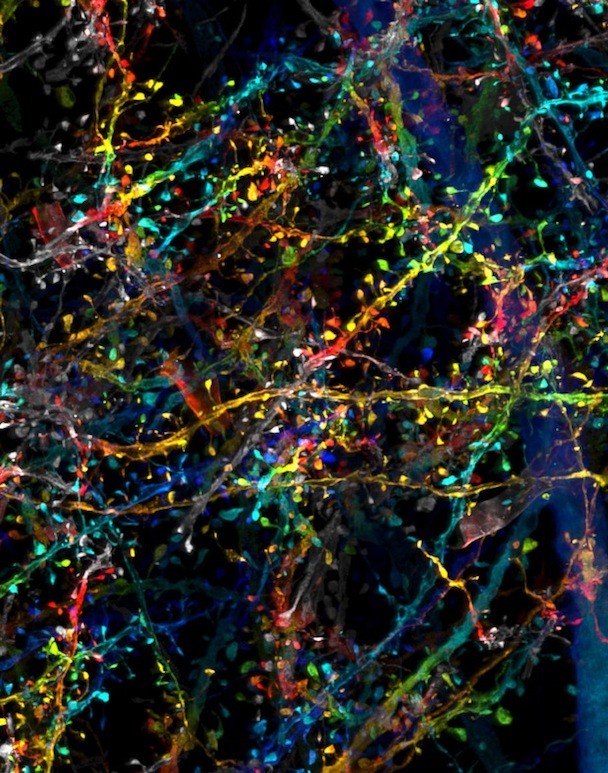
Scientists Ruixuan Gao, PhD, and Shoh Asano, PhD, of MIT McGovern Institute for Brain Research and Media Lab, first had the idea to combine expansion microscopy with lattice light-sheet microscopy.
Previously, expansion microscopy had only been used to image single cells and thin sections of tissues. It works by expanding a sample symmetrically across three dimensions. “This preserves the relative spatial distances between cellular and molecular structures,” explains neuroscientist Chinfei Chen, MD, PhD, who was not part of the study.
To accomplish the expansion, the researchers embedded tissues with polymers — similar to the ones found in diapers — that are able to swell to many times their original size. Expanding the tissue allows scientists to image with much greater resolution and accuracy, revealing structures that were previously indistinguishable.
Most prior imaging methods shine light through the entirety of a sample while recording only a small section. This extended exposure can damage fluorescent proteins or chemical tags in the tissue, making mapping a large area nearly impossible. Lattice light-sheet microscopy, developed by Betzig, shines a thin, gentle beam of light through only the plane being imaged. This minimizes damage to the rest of the sample. Computers then stitch together the millions of images produced into one final product.
Crunching data
One of the biggest challenges for the team was processing the massive amount of data they recorded. Software that traditionally handled a few megabytes had to be adapted to process terabytes at a time. In total, the images from the project used roughly 45,000 traditional DVDs’ worth of data.
By combining these two methods, the team was able to capture an image of a fly brain that would have taken decades previously in less than a week.
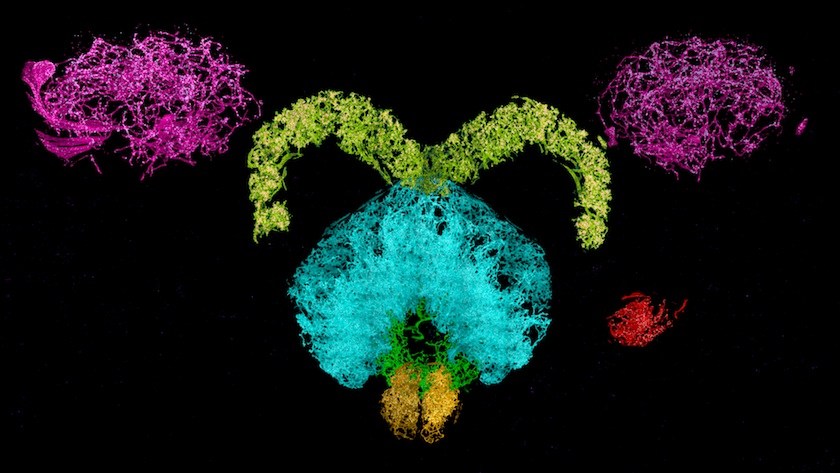
The 3D images from the new technique have nanoscale resolution, revealing minute sub-cellular anatomy — such as the location and detailed morphology of dendritic spines, critical for learning and development. Researchers can tag and trace specific proteins through the brain, allowing them to explore new questions about physiology and pathology. The team has already used the new technology to trace connections across the brain and quantify how many neurons release dopamine.
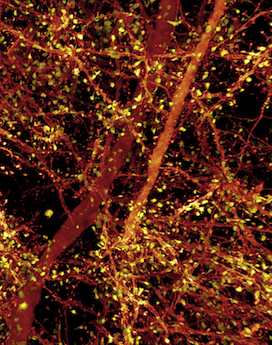
“The visualization of brain tissue at the resolution and spatial depth described in this paper is truly remarkable,” says Chen, who directs the imaging core at Boston Children’s Intellectual and Developmental Disabilities Research Center (IDDRC). “It allows neuroscientists to see subcellular ultrastructure in the context of the neuronal network. I would certainly explore the possibility of incorporating this technology in the imaging core.”
Bringing neuroscience into the future
Upadhyayula is part of a team, led by Betzig, that is now building one of seven next-generation, multi-functional microscopes designed to leverage adaptive optics — a tool originally developed by astronomers for viewing distant stars through the interference of the atmosphere. This would create a microscope more versatile and capable than any currently in existence, and is expected to be online in the Kirchhausen Lab at HMS and Boston Children’s early spring 2019.
While every brain has underlying similarities, individuals differ. How? That’s a question the new system is well positioned to answer. It could also be used to compare healthy brains to diseased brains, young brains to old brains and male brains to female brains. It could even be used to look at non-nervous tissue.
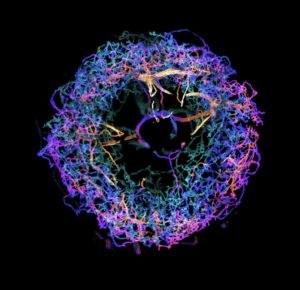
Upadhyayula believes that the team’s work overcomes the current barriers in scalable super-resolution imaging of tissues. “This technology ushers in a new era for neuroscience,” he says.
The work was funded by HHMI, John Doerr, the Open Philanthropy Project, the National Institutes of Health, the Intelligence Advanced Research Projects Activity (IARPA), the U. S. Army Research Laboratory and the U.S. Army Research Office, the U.S.-Israel Binational Science Foundation, Biogen and Ionis Pharmaceuticals.
Ruixuan Gao, PHD, of MIT and HHMI, and Shoh Asano, PhD, of MIT are co-first authors on the paper with Upadhyayula. Lead researcher Eric Betzig is now an HHMI investigator at UC Berkeley. See the paper for a full list of authors and grants.
More on the technology is available on the HHMI and HMS websites. Read about how members of the team used adaptive optics to look inside living organisms.
Related Posts :
-

The thalamus: A potential therapeutic target for neurodevelopmental disorders
Years ago, as a neurology resident, Chinfei Chen, MD, PhD, cared for a 20-year-old woman who had experienced a very ...
-

Could peripheral neuropathy be stopped before it starts?
An increase in high-fat, high-fructose foods in people’s diets has contributed to a dramatic increase in type 2 diabetes. This, ...
-

A new tool could exponentially expand our understanding of bacteria
How do bacteria — harmless ones living in our bodies, or those that cause disease — organize their activities? A new study, ...
-

AI-enabled medical devices are burgeoning, but many haven’t been tested in children
Medical devices that incorporate artificial intelligence and machine learning are proliferating. In 2013, the FDA approved fewer than 10 such devices; by 2023, ...